Using_the_SpinyRiceFlowersPackage
Using_the_SpinyRiceFlowersPackage.RmdThe package depends on a number of R packages. To install a package, say “ggplot2”, the following commands can be used.
if(!require('ggplot2')) {
install.packages('ggplot2')
library('ggplot2')
}
#> Loading required package: ggplot2
if(!require('ggdensity')) {
install.packages('ggdensity')
library('ggdensity')
}
#> Loading required package: ggdensity
if(!require('dplyr')) {
install.packages('dplyr')
library('dplyr')
}
#> Loading required package: dplyr
#> Warning: package 'dplyr' was built under R version 4.4.2
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(SpinyRiceFlowers)Installation of the Package
Choose a directory you want to work in and save the zip file to that directory. Then use the following commands.
install.packages("SpinyRiceFlowers_0.1.2.zip")
library(SpinyRiceFlowers)Datasets
The package includes two types of data sets, the longitudinal data sets and the Statewide evaluation data sets.
The longitudanal data sets give the the status of plants collected annually from 2017 to 2021. The data sets are:
| BonThomas A | PimeleaA |
| BonThomas B | PimeleaB |
| DentonAveA | PimeleaC |
| DentonAveB | PimeleaD |
| IramooA | PimeleaE |
| IramooB | PimeleaF |
To print out the data set, just type the name. Alternatively, you can
get the first few rows with the head()command.
head(PimeleaA, n=10)
#> # A tibble: 10 × 8
#> `Tag ID` `Status 2017` `Status 2018` `Status 2019` `Status 2020`
#> <dbl> <chr> <chr> <chr> <chr>
#> 1 78 Existing Alive Alive Alive
#> 2 79 Existing Alive Alive Alive
#> 3 82 Existing Alive Alive Alive
#> 4 80 Existing Alive Alive Alive
#> 5 83 Existing Alive Alive Alive
#> 6 429 Germinant Alive Alive Alive
#> 7 421 Germinant Alive Alive Alive
#> 8 430 Germinant Alive Alive Alive
#> 9 422 NA Germinant Alive Alive
#> 10 423 NA Germinant Alive Alive
#> # ℹ 3 more variables: `Status 2021` <chr>, `x-axis` <dbl>, `y-axis` <dbl>The Statewide evaluation data sets are:
| Site19 | Site29 |
| Site22 | Site45 |
For convenience, a number of derived data sets related to BonThomasA have been included in the package:
| AllPlantsby2020 |
| BonThomasADied2021 |
| BonThomasARecruit2021 |
Information about the datsets can be obtained using the help command.
help(AllPlantsby2020)Heatmaps
Heatmaps can be prepared using the HeatMap() command.
Required fields include the co-ordinates of the plants, optionally the
number of plants at that co-ordinate (called the weights), the
and
ranges, and a title.
HeatMap(AllPlantsby2020[,2:3],xrange=c(0,35), yrange=c(0,15), gridsep=5, title="Heat map for Bon Thomas A")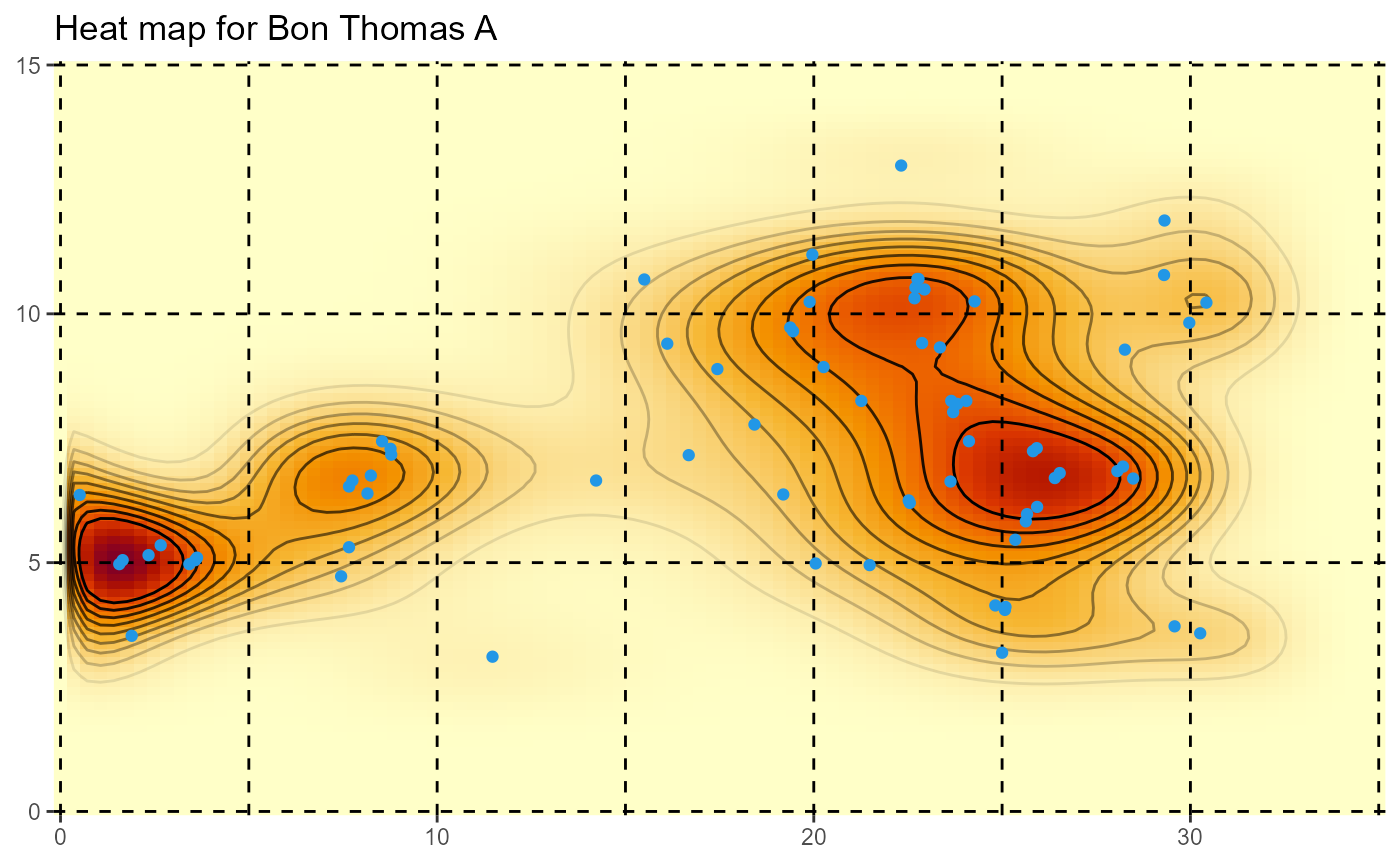
For the State-wide evaluation data, the locations are actually the mid-points of 1m by 1m quadrats, and the weights are the corresponding number of plants in the quadrats. The area of the plotted points is also proprtional to the number of plants.
HeatMap(Site22[,2:3], weights=Site22$NUMPimelea,xrange=c(10,291), yrange=c(1,19), gridsep=10,title="Heat map for Site 22")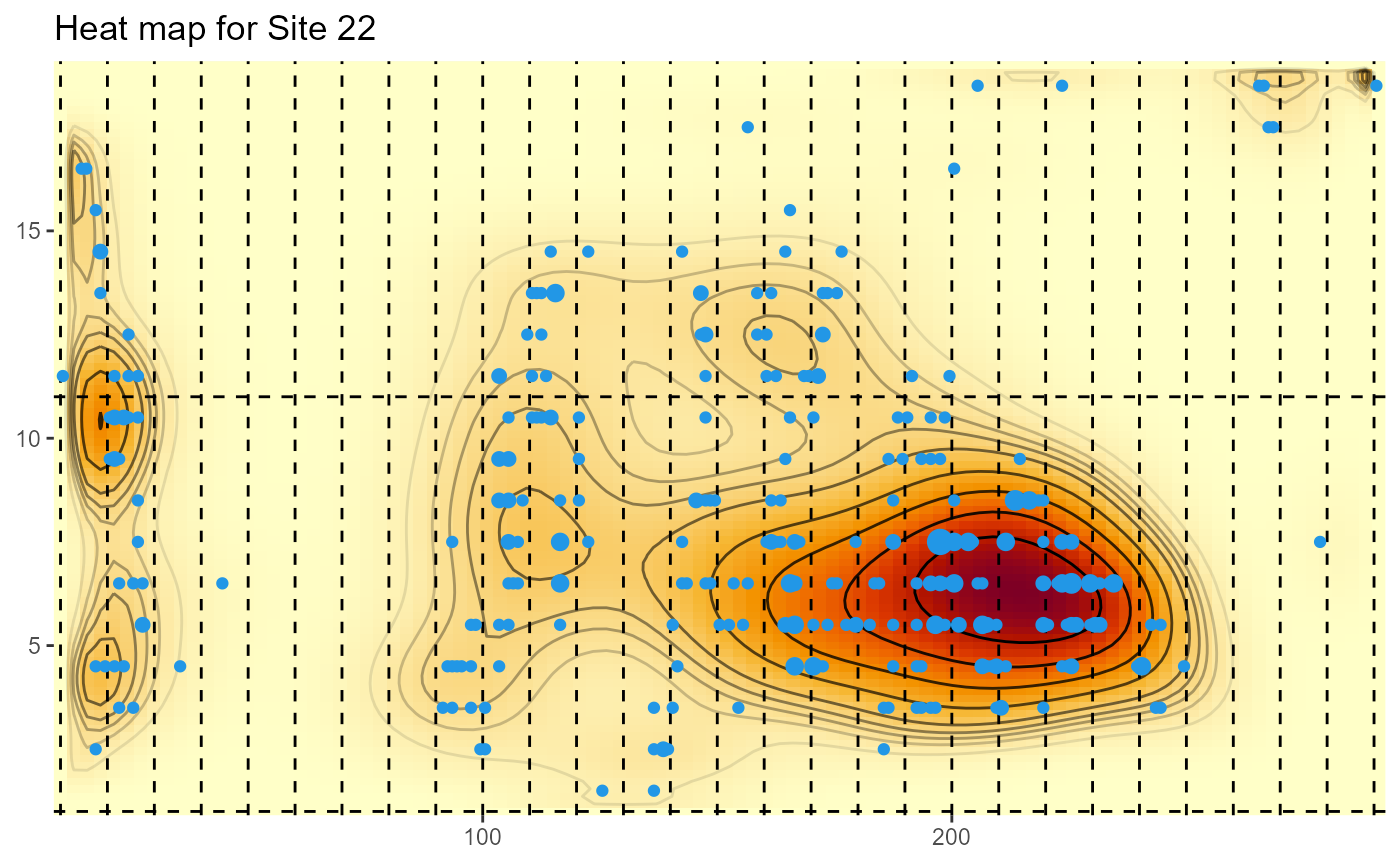
HeatMap(Site29[,2:3], weights=Site29$NUMPimelea,xrange=c(11,25), yrange=c(0,11), gridsep=5,title="Heat map for Site 29")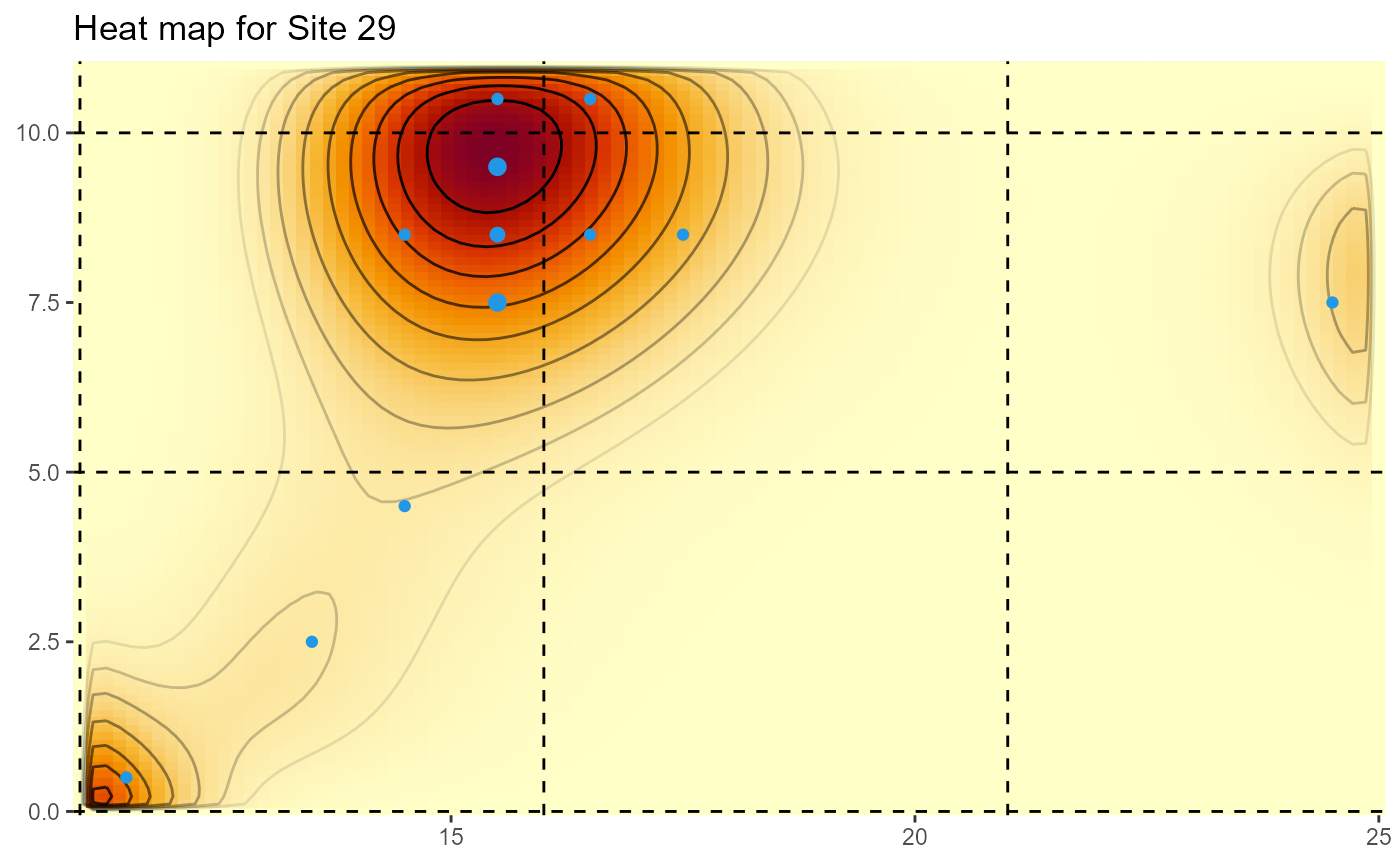
HeatMap(Site45[,2:3], weights=Site45$NUMPimelea,xrange=c(11,86), yrange=c(11,263), gridsep=20,title="Heat map for Site 45")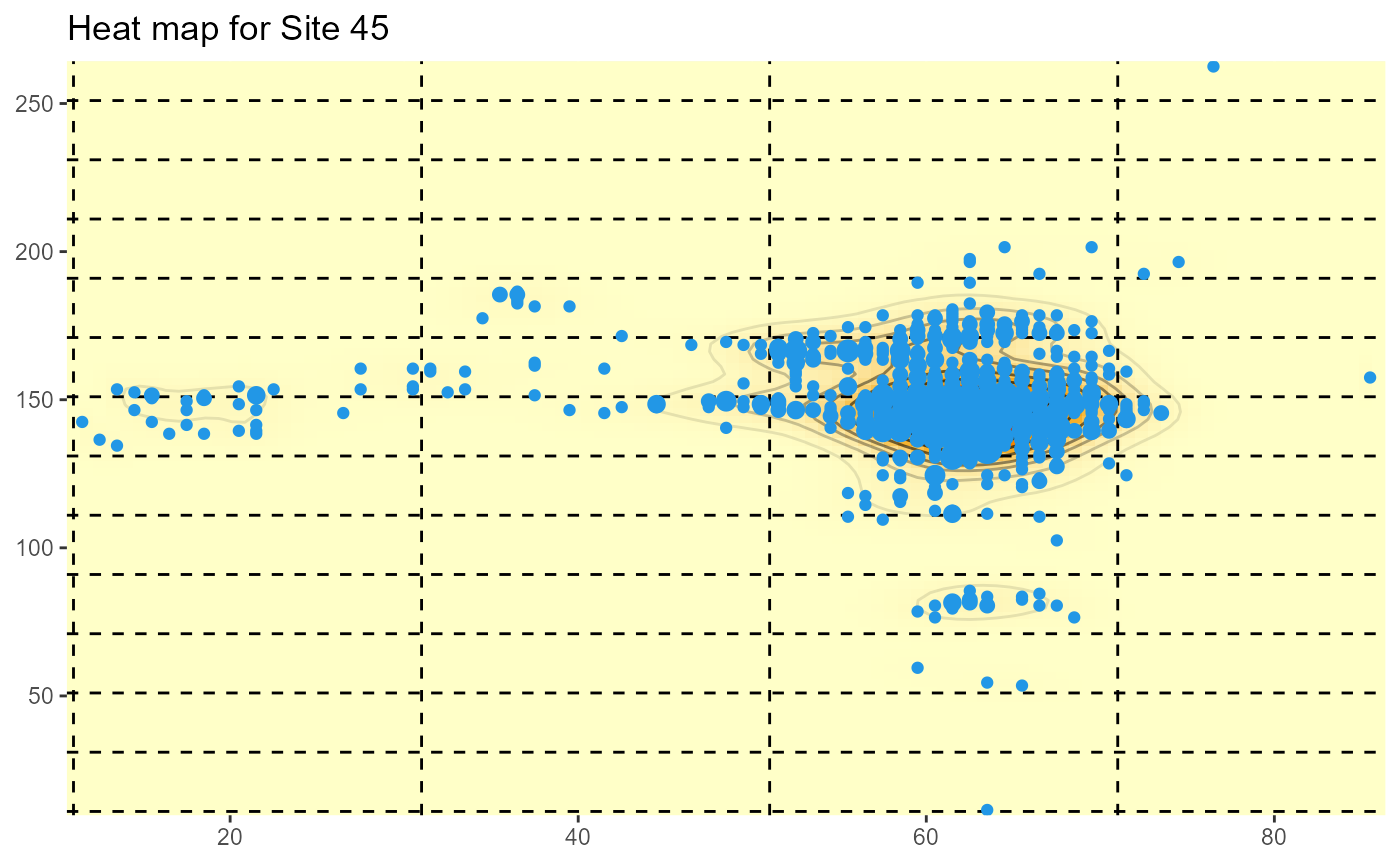
Quadrat Probabilities
Quadrat probabilities can be calculated using the
quadratprobs() function. Required inputs are the locations
of plants, the lower left coordinate of the grid, with default (0,0),
the size of the quadrats, and the minimum grid values, and the maximum
grid values. In the example below, it is not necessary to specify the
quadratstart argument aswith 5m by 5m quadrats, 5 divides
35 and 15 evenly.
qprobs5 <- quadratprobs(AllPlantsby2020[,2:3], quadratstart=c(0,0),quadratsize=5,
xmin=c(0,0),xmax=c(35,15) )
knitr::kable(qprobs5, digits=3)| [0,5) | [5,10) | [10,15) | [15,20) | [20,25) | [25,30) | [30,35) | |
|---|---|---|---|---|---|---|---|
| [10,15) | 0.000 | 0.000 | 0.008 | 0.036 | 0.067 | 0.041 | 0.019 |
| [5,10) | 0.067 | 0.080 | 0.047 | 0.087 | 0.172 | 0.153 | 0.026 |
| [0,5) | 0.056 | 0.021 | 0.011 | 0.010 | 0.036 | 0.045 | 0.015 |
On the other hand, for 4m by 4m quadrats, it is useful to be able to
specify the quadratstart argument. Note that only internal
quadrats are included in the output of quadratprobs().
qprobs4 <- quadratprobs(AllPlantsby2020[,2:3], quadratstart=c(2,1),quadratsize=4,
xmin=c(0,0),xmax=c(35,15) )
knitr::kable(qprobs4, digits=3)| [2,6) | [6,10) | [10,14) | [14,18) | [18,22) | [22,26) | [26,30) | [30,34) | |
|---|---|---|---|---|---|---|---|---|
| [9,13) | 0.000 | 0.001 | 0.008 | 0.033 | 0.071 | 0.076 | 0.046 | 0.027 |
| [5,9) | 0.051 | 0.065 | 0.033 | 0.039 | 0.076 | 0.123 | 0.098 | 0.018 |
| [1,5) | 0.034 | 0.016 | 0.010 | 0.006 | 0.015 | 0.037 | 0.034 | 0.015 |
Inclusion Probabilities
Once the quadrat probabilities are calculated, the
inclusionprobs() function can calculate the inclusion
probabilities. The following gives the inclusion probabilities for
various sample sizes. The quadrats are ordered by column of the quadrat
probabilities matrix with the top-left quadrat labelled
q1.
incprobs <- inclusionprobs(qprobs5,nsim=10000)
knitr::kable(incprobs, digits=3)| ss | q1 | q2 | q3 | q4 | q5 | q6 | q7 | q8 | q9 | q10 | q11 | q12 | q13 | q14 | q15 | q16 | q17 | q18 | q19 | q20 | q21 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.0000 | 0.0673 | 0.0580 | 0.0001 | 0.0775 | 0.0190 | 0.0095 | 0.0497 | 0.0122 | 0.0342 | 0.0828 | 0.0091 | 0.0716 | 0.1667 | 0.0358 | 0.0449 | 0.1570 | 0.0451 | 0.0199 | 0.0255 | 0.0141 |
| 2 | 0.0000 | 0.1333 | 0.1199 | 0.0002 | 0.1602 | 0.0411 | 0.0171 | 0.1020 | 0.0252 | 0.0765 | 0.1654 | 0.0198 | 0.1398 | 0.3282 | 0.0742 | 0.0867 | 0.2971 | 0.0935 | 0.0403 | 0.0505 | 0.0290 |
| 3 | 0.0000 | 0.2051 | 0.1812 | 0.0006 | 0.2419 | 0.0658 | 0.0250 | 0.1527 | 0.0384 | 0.1196 | 0.2588 | 0.0327 | 0.2107 | 0.4685 | 0.1165 | 0.1290 | 0.4223 | 0.1406 | 0.0632 | 0.0791 | 0.0483 |
| 4 | 0.0001 | 0.2785 | 0.2398 | 0.0022 | 0.3274 | 0.0922 | 0.0330 | 0.2067 | 0.0513 | 0.1674 | 0.3470 | 0.0452 | 0.2834 | 0.5853 | 0.1599 | 0.1785 | 0.5421 | 0.1914 | 0.0878 | 0.1130 | 0.0678 |
| 5 | 0.0001 | 0.3490 | 0.3053 | 0.0027 | 0.4119 | 0.1198 | 0.0458 | 0.2654 | 0.0674 | 0.2128 | 0.4381 | 0.0596 | 0.3561 | 0.6831 | 0.2048 | 0.2333 | 0.6422 | 0.2480 | 0.1176 | 0.1468 | 0.0902 |
| 6 | 0.0001 | 0.4272 | 0.3747 | 0.0033 | 0.4872 | 0.1522 | 0.0580 | 0.3238 | 0.0863 | 0.2625 | 0.5222 | 0.0758 | 0.4289 | 0.7653 | 0.2526 | 0.2905 | 0.7327 | 0.3071 | 0.1488 | 0.1841 | 0.1167 |
| 7 | 0.0001 | 0.5054 | 0.4404 | 0.0038 | 0.5672 | 0.1845 | 0.0722 | 0.3866 | 0.1063 | 0.3137 | 0.6007 | 0.0929 | 0.5093 | 0.8340 | 0.3060 | 0.3493 | 0.8048 | 0.3691 | 0.1838 | 0.2253 | 0.1446 |
| 8 | 0.0001 | 0.5855 | 0.5081 | 0.0045 | 0.6435 | 0.2243 | 0.0902 | 0.4564 | 0.1297 | 0.3705 | 0.6749 | 0.1111 | 0.5838 | 0.8839 | 0.3655 | 0.4091 | 0.8584 | 0.4329 | 0.2234 | 0.2705 | 0.1737 |
| 9 | 0.0001 | 0.6588 | 0.5758 | 0.0052 | 0.7184 | 0.2680 | 0.1078 | 0.5251 | 0.1541 | 0.4362 | 0.7411 | 0.1356 | 0.6553 | 0.9245 | 0.4282 | 0.4725 | 0.9051 | 0.5024 | 0.2593 | 0.3175 | 0.2090 |
| 10 | 0.0001 | 0.7278 | 0.6517 | 0.0065 | 0.7809 | 0.3168 | 0.1315 | 0.5887 | 0.1818 | 0.4995 | 0.8040 | 0.1607 | 0.7206 | 0.9522 | 0.4970 | 0.5410 | 0.9385 | 0.5757 | 0.3080 | 0.3718 | 0.2452 |
| 11 | 0.0001 | 0.7868 | 0.7203 | 0.0077 | 0.8375 | 0.3685 | 0.1565 | 0.6619 | 0.2164 | 0.5667 | 0.8582 | 0.1882 | 0.7859 | 0.9713 | 0.5652 | 0.6128 | 0.9637 | 0.6476 | 0.3642 | 0.4316 | 0.2889 |
| 12 | 0.0002 | 0.8423 | 0.7837 | 0.0089 | 0.8876 | 0.4271 | 0.1884 | 0.7297 | 0.2557 | 0.6398 | 0.9039 | 0.2210 | 0.8446 | 0.9845 | 0.6404 | 0.6850 | 0.9817 | 0.7186 | 0.4210 | 0.4985 | 0.3374 |
| 13 | 0.0002 | 0.8951 | 0.8448 | 0.0113 | 0.9286 | 0.4969 | 0.2252 | 0.7953 | 0.3045 | 0.7119 | 0.9412 | 0.2632 | 0.8959 | 0.9919 | 0.7093 | 0.7580 | 0.9899 | 0.7857 | 0.4868 | 0.5730 | 0.3913 |
| 14 | 0.0002 | 0.9355 | 0.8976 | 0.0142 | 0.9570 | 0.5735 | 0.2697 | 0.8599 | 0.3669 | 0.7859 | 0.9656 | 0.3129 | 0.9360 | 0.9958 | 0.7859 | 0.8276 | 0.9962 | 0.8506 | 0.5586 | 0.6541 | 0.4563 |
| 15 | 0.0002 | 0.9652 | 0.9382 | 0.0170 | 0.9790 | 0.6633 | 0.3262 | 0.9128 | 0.4446 | 0.8536 | 0.9847 | 0.3808 | 0.9648 | 0.9986 | 0.8555 | 0.8890 | 0.9985 | 0.9063 | 0.6452 | 0.7376 | 0.5389 |
| 16 | 0.0004 | 0.9848 | 0.9695 | 0.0223 | 0.9917 | 0.7566 | 0.4061 | 0.9531 | 0.5372 | 0.9148 | 0.9939 | 0.4702 | 0.9823 | 0.9997 | 0.9159 | 0.9417 | 0.9995 | 0.9491 | 0.7422 | 0.8231 | 0.6459 |
| 17 | 0.0005 | 0.9957 | 0.9883 | 0.0284 | 0.9974 | 0.8575 | 0.5172 | 0.9810 | 0.6631 | 0.9589 | 0.9987 | 0.5888 | 0.9951 | 1.0000 | 0.9574 | 0.9770 | 0.9999 | 0.9785 | 0.8460 | 0.9051 | 0.7655 |
| 18 | 0.0009 | 0.9985 | 0.9971 | 0.0419 | 0.9997 | 0.9468 | 0.6850 | 0.9937 | 0.8218 | 0.9894 | 0.9996 | 0.7574 | 0.9988 | 1.0000 | 0.9866 | 0.9937 | 1.0000 | 0.9950 | 0.9369 | 0.9660 | 0.8912 |
| 19 | 0.0013 | 0.9999 | 1.0000 | 0.0707 | 1.0000 | 0.9974 | 0.9752 | 0.9999 | 0.9872 | 0.9994 | 1.0000 | 0.9812 | 1.0000 | 1.0000 | 0.9998 | 0.9999 | 1.0000 | 0.9999 | 0.9957 | 0.9988 | 0.9937 |
| 20 | 0.0080 | 1.0000 | 1.0000 | 0.9920 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 21 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
Number of Plants Alive at a particular year
(NumberAliveBonThomasA2020 <- NumberAlive(BonThomasA, 2020))
#> Alive
#> 66Quadrat Counts of Changes
Change(BonThomasA,2021, quadratsize=5)
#> x
#> y [0,5) [5,10) [10,15) [15,20) [20,25) [25,30) [30,35]
#> [10,15] 0 0 0 0 1 0 0
#> [5,10) -1 0 0 0 1 0 0
#> [0,5) 1 0 0 0 -1 0 0Estimated Total
The EstimatedTotal() command gives the mean, standard
deviation, and coefficient of variation (%) for a specified sample
size.
EstimatedTotal(BonThomasA, 2021, ss=5, probs=qprobs5, incprobs=incprobs[5,-1])
#> [[1]]
#> Alive
#> 66.94107
#>
#> [[2]]
#> [1] 3.33128
#>
#> [[3]]
#> Alive
#> 4.98
EstimatedTotal(BonThomasA, 2021, ss=10, probs=qprobs5, incprobs=incprobs[10,-1])
#> [[1]]
#> Alive
#> 67.00685
#>
#> [[2]]
#> [1] 1.587954
#>
#> [[3]]
#> Alive
#> 2.37
EstimatedTotal(BonThomasA, 2021, ss=15, probs=qprobs5, incprobs=incprobs[15,-1])
#> [[1]]
#> Alive
#> 67.00507
#>
#> [[2]]
#> [1] 0.5580785
#>
#> [[3]]
#> Alive
#> 0.83